Introduction
On Nov. 24, 2021, the variant B.1.1.529 was first identified in South Africa, and only two days later, WHO announced that B.1.1.529 constitutes a variant of concern, named “Omicron”. There are more than 30 mutation sites on the S gene of variant B.1.1.529. Omicron has a greater binding affinity than the original SARS-CoV-2 virus, with levels more comparable to what we see with the Delta variant. Some of the mutations have been detected in previous variants, such as Alpha and Delta, and have been associated with increased transmissibility and immune evasion. Many of the other identified mutations are not yet well characterized and have not been identified in other currently circulating variants. More investigations are underway to determine the possible impact of these mutations. Omicron includes Pango lineage B.1.1.529 and descendent Pango lineages BA.1, BA.1.1, BA.2 and BA.3.
Features
- Identify Omicron variant by simultaneous detection of ORF1ab, and four specific mutation sites of S gene
- Automated result interpretation add-in for STC-96A & 96A Plus*
- Semi-automated interpretation tool available for other real-time PCR instruments
- Endogenous internal control (IC) ensures the whole process is reliable
Automated Result Interpretation for Mutation
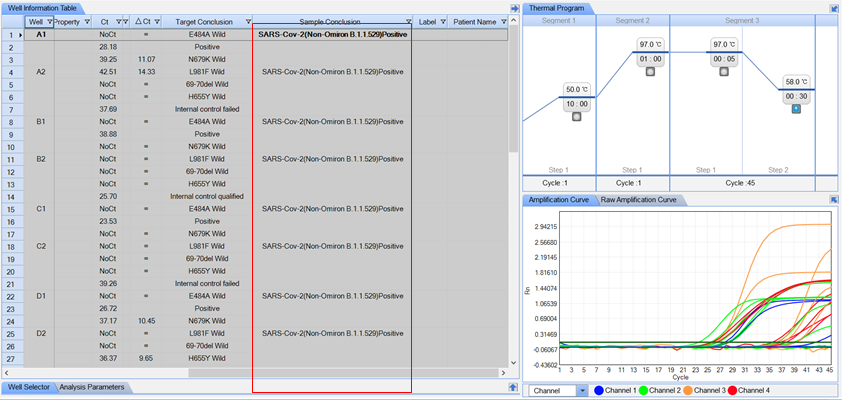
Parameters
| Target |
ORF1ab, mutations E484A, N679K, L981F, and H655Y of S gene |
| Coincidence with an internal positive reference |
100% |
| Coincidence with an internal negative reference |
100% |
| Limit of detection |
500 copies/mL |
| Sample type |
Nasopharyngeal swabs, throat swabs, sputum specimens. |
Certificate
For Research Use Only. Waiting for the clearance of regulatory.
